β-内酰胺酶的产生、进化与传播是革兰阴性菌对β-内酰胺类药物耐药的最主要机制[1-2]。根据不同底物谱可将β-内酰胺酶分为青霉素酶、头孢菌素酶和碳青霉烯酶。碳青霉烯酶是一类能够对碳青霉烯类药物产生水解作用的β-内酰胺酶,主要包括Ambler分子结构分类法中的A类、B类、D类,其中A类酶和D类酶利用丝氨酸残基发挥水解作用,按功能分别属于Bush分类法中的2f和2d亚组;而B类酶利用金属离子才具有活性,属于Bush分类法中第3组[3](见表 1);D类β-内酰胺酶有14个亚家族,其中OXA型种类最为丰富,并不断发现新的变体[4]。截至2022年9月,已有1 144种OXA酶被β-内酰胺酶数据库(http://www.bldb.eu/BLDB.php?prot=D#OXA)收录,其中水解碳青霉烯类药物的OXA型酶数量增加较多,给全球公共卫生带来严重威胁。
| 表 1 碳青霉烯酶分类及功能特点 |
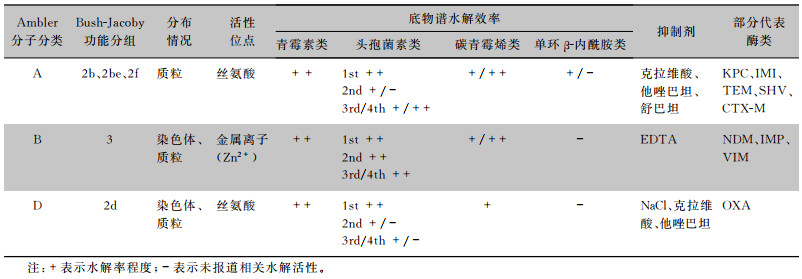
|
苯唑西林酶(oxacillinase, OXA酶)是最早报道的β-内酰胺酶之一,因其对苯唑西林和氯唑西林的水解率高于青霉素而命名[5]。早期发现的OXA酶仅对青霉素类药物有活性,随着变体的不断出现,其底物谱不断扩大,部分OXA酶变体不仅表现出对头孢菌素类抗生素具有水解能力,甚至对碳青霉烯类抗生素也产生一定的水解活性,其活性不受克拉维酸、他唑巴坦、舒巴坦等酶抑制剂的抑制[6]。
1976年,Sykes等[7]通过基因序列分析首次发现并命名OXA酶:OXA-1~OXA-3,并以其共有的保守序列作为后续OXA酶分类的基础,此后根据发现时间进行数字编号命名OXA酶(OXA酶的种类及分类见表 2)。随着OXA酶家族成员的增加,BLDB数据库将氨基酸同源性高、底物谱相似的酶以首个命名的OXA酶的亚家族来命名,称为OXA-X-like酶,如OXA-48-like酶、OXA-23-like酶。OXA酶根据来源不同可分为获得性及内源性两大类,blaOXA基因既可由质粒携带也可由染色体携带[5]。鲍曼不动杆菌和铜绿假单胞菌中染色体携带blaOXA基因较多,肺炎克雷伯菌中质粒携带则更为常见。近年来,全球范围内陆续报道了革兰阴性肠杆菌中具有碳青霉烯酶活性的新型OXA酶,这显示细菌耐药形势日益严峻。
| 表 2 OXA型β-内酰胺酶种类及来源 |
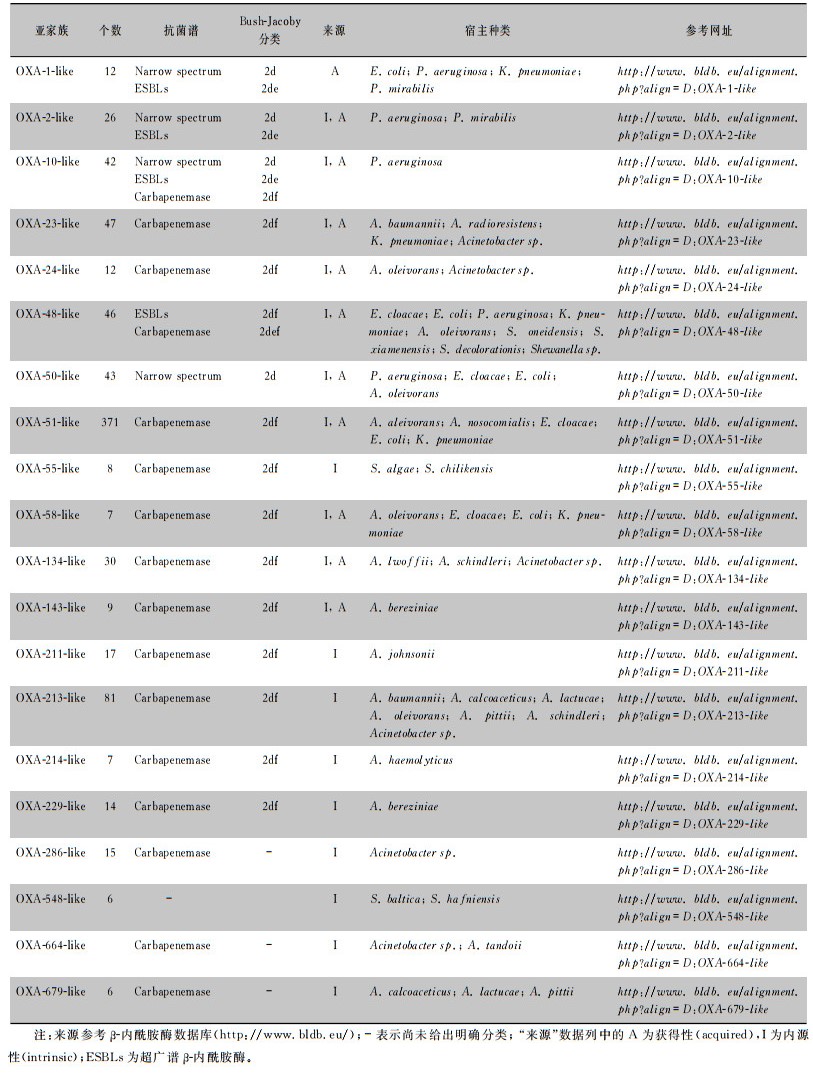
|
目前,OXA酶宿主菌范围不断扩大,从最初发现的鲍曼不动杆菌、铜绿假单胞菌,到目前发现的肺炎克雷伯菌、大肠埃希菌、阴沟肠杆菌以及其他肠杆菌[8-10]。
不动杆菌中几乎所有OXA酶都具有碳青霉烯酶活性,联合外排泵过表达机制可介导对碳青霉烯类抗生素高水平耐药。中东和北非等地区耐碳青霉烯类鲍曼不动杆菌(CRAB)分离率超过70%[11]。鲍曼不动杆菌携带染色体固有OXA-51-like酶,该家族已发现363个变种,是造成鲍曼不动杆菌耐药的首要因素。OXA-23是鲍曼不动杆菌中常见的获得性OXA酶,最初在苏格兰发现的CRAB菌株中分离并命名,现已报道发现44个变体,主要在欧美地区流行[12-14]。OXA-24/40首次在西班牙分离出的CRAB中发现,此后陆续衍生出11个变体,在中国、美国、埃及、土耳其及伊朗等国家均有流行[15-17]。2003年在法国图卢兹发现的1株CRAB中检测出OXA-58,其与其他OXA家族同源性低,被列为OXA新的亚家族,该家族现已衍生出7个变种,主要通过质粒传播,在欧洲、亚洲及拉美地区引起医院感染暴发[18-20]。
OXA-48-like酶由于主要在肠杆菌目细菌中传播而备受关注。自2001年在土耳其伊斯坦布尔分离的肺炎克雷伯菌中首次检出水解碳青霉烯类活性的OXA-48以来,其变体在欧洲、亚洲、北非、中东地区的肠杆菌目细菌中广泛传播[21-22]。截至2022年6月,世界范围内已确认和命名45个OXA-48-like酶。我国主要以OXA-181及OXA-232两大变体为主,两者仅存在1个氨基酸差异。自印度1株肺炎克雷伯菌分离出OXA-181后,在其他肠杆菌中也被分离出来,呈现出世界范围内流行的态势[23-25]。2013年,法国1株肺炎克雷伯菌中首次检出OXA-232,而后在我国江浙沪地区相继检出,主要通过质粒进行菌株间水平转移[26-28]。OXA-50-like酶为假单胞菌所特有,现已衍生出43个变体,其与外排泵过表达、外膜蛋白减少或丢失等机制共同作用,是造成假单胞菌多重耐药的重要原因[29]。Nitz等[30]首次在临床分离的铜绿假单胞菌中检测出OXA-23和OXA-51,该菌株存在高水平多重耐药。变形杆菌具有携带多种类型水解碳青霉烯类OXA酶的特征。Bonnin等[31]发现OXA-23、OXA-24及OXA-58在法国和比利时地区分离的变形杆菌中流行。我国也在奇异变形杆菌中检出了blaOXA-23和blaOXA-48基因[32]。
转座子、整合子等可移动元件介导的耐药基因的水平转移是导致耐药基因在不同种属之间传播的重要机制[33]。此外,插入元件中的调控序列能够促进基因的过表达,例如鲍曼不动杆菌中常见的插入序列ISAba1,隶属于IS4家族,能够提供强启动子以增强其下游blaOXA-23、blaOXA-27等基因的表达,也可形成转座子介导blaOXA基因的转移。接合型质粒携带的基因可随质粒转移,更易造成耐药性的传播[34]。
3 OXA酶功能研究不同OXA酶具有不同的底物谱,Bush-Jacoby分类法[35]根据酶的功能和表型,将OXA酶分为2d、2de、2df和2def四个功能组。OXA酶对氯唑西林或苯唑西林水解活性高于对青霉素的50%,则被归为2d功能组,其中大部分属于窄谱酶(OXA-1-like酶、OXA-50-like酶),其仅对青霉素类药物有明显的水解活性,对头孢菌素类抗生素作用微弱,且活性不会被克拉维酸抑制。近年来,部分窄谱酶因单一或多个氨基酸位点突变而发生底物谱变化,使得某些OXA酶具有广谱性质,被归为2de亚组ESBLs,主要为OXA-2-like酶及OXA-10-like酶。土耳其分离的1株铜绿假单胞菌中发现的OXA-15是由OXA-2发生单一氨基酸突变形成的超广谱OXA酶,对头孢他啶有较高的水解活性。OXA-10-like酶中具有广谱水解活性的变体较多。与OXA-10相比,OXA-11有两个氨基酸差异,增加了其对三代头孢的水解能力。OXA-145是OXA-10的另一个变体,第165位氨基酸的缺失扩大该酶家族的底物谱,减弱了对青霉素类药物的水解活性。Bonnin等[36]在法国1株铜绿假单胞菌中检出OXA-198,被归为一个新的D类酶亚组,blaOXA-198位于IncP型质粒携带的Ⅰ类整合子上,显著降低对碳青霉烯类抗生素的敏感性。Kotsakis等[37]在医院污水分离的肠杆菌中发现同样定位于Ⅰ类整合子上的blaOXA-655和blaOXA-656,均为OXA-10的新变体,这些变体具有更强的碳青霉烯类水解能力,存在水平传播的风险。
目前,关注较多的是由于活性位点变化而产生碳青霉烯类水解能力的2df亚组OXA酶。OXA-23是第一个被确认水解碳青霉烯类的OXA酶,其对亚胺培南的水解活性显著提高,而对广谱头孢菌素类抗生素及氨曲南水解能力弱[38]。与OXA-23相比,OXA-27有两个氨基酸差异,同样表现出对碳青霉烯类抗生素的水解活性。此外,我国报道了单个氨基酸突变产生的新变种OXA-423,同样具有碳青霉烯类抗生素的水解活性,但对克拉维酸、他唑巴坦等酶抑制剂敏感[39]。blaOXA-51基因编码一种弱的碳青霉烯酶,多为内源性携带,也可由质粒携带,质粒相邻的ISAba1可促使该基因过表达,表现出对碳青霉烯类药物的高水平耐药[40]。OXA-58由质粒携带,对青霉素类和碳青霉烯类抗生素水解活性较弱,而对广谱头孢菌素类抗生素活性较差,多个ISs协同转座可能是其传播的重要方式[41]。
肠杆菌中最常见的碳青霉烯酶为OXA-48,IS1999及多种质粒的高效转移加速了blaOXA-48基因在细菌间的水平传播[42]。OXA-181是分布最为广泛的OXA-48-like酶家族成员,其与OXA-48有四个氨基酸差异,且表现出极为相似的水解特征。Aertker等[43]证明单一氨基酸替换能够使OXA-48-like酶对碳青霉烯类抗生素表现出不同的水解活性。OXA-162与OXA-48仅有一个氨基酸差异,同样能够介导肠杆菌对碳青霉烯类抗生素耐药。Sommer等[44]在大肠埃希菌中发现质粒携带的blaOXA-484基因,OXA-484与OXA-181存在1个氨基酸替换,其对碳青霉烯类抗生素的水解能力略低于OXA-48及OXA-181。比利时分离出的1株肺炎克雷伯菌中发现了OXA-519,其与OXA-48有1个氨基酸差异,表现出对哌拉西林/他唑巴坦耐药和对亚胺培南敏感性降低[45]。然而,并不是所有的OXA-48变体都对碳青霉烯类抗生素有水解活性。OXA-405与OXA-48具有相似的主体结构,blaOXA-405位于转座子Tn1999上,IS1R插入替代了保守序列缺失的4个氨基酸,使得OXA-405对碳青霉烯类几乎完全失去水解活性,仅表现出ESBL活性[46]。Poirel等[47]在肠杆菌中发现OXA-163,与OXA-48相比有5个氨基酸变化,能够水解青霉素类、第三代头孢菌素类以及碳青霉烯类抗生素,被分类为兼具头孢菌素类和碳青霉烯类抗生素水解活性的2def亚组。Gomez等[48]从阿根廷1例白血病患者体内分离出携带OXA-163的肺炎克雷伯菌,治疗后又在其体内分离出携带OXA-247的肺炎克雷伯菌,OXA-247是OXA-163的新变体,两者有相似的底物谱,但对广谱头孢菌素类抗生素的水解活性减弱。
20世纪70年代β-内酰胺酶抑制剂应用于临床,常见的酶抑制剂包括克拉维酸、他唑巴坦和舒巴坦等,能够对OXA酶产生不同程度的抑制作用,他唑巴坦对OXA-1仅表现弱抑制作用;对于同一家族成员,他唑巴坦对OXA-32表现强抑制作用,克拉维酸则相对较弱,而OXA-53对后者更加敏感[49]。研究[50-51]显示,头孢他啶/阿维巴坦、亚胺培南/西司他丁等新型酶抑制剂复合制剂对某些产D类酶的肠杆菌有一定杀灭作用。
4 OXA酶与希瓦氏菌的关系研究希瓦氏菌是一种水体环境生存的革兰阴性菌,早期认为该类菌仅存在于腐败变质的高蛋白水产食品中。近年研究[52-53]显示,希瓦氏菌的部分种属与人类感染密切相关,易感人群可通过接触或食用含有希瓦氏菌的水、海产、食品等引起腹泻、溶血性反应以及软组织感染等疾病。临床常见的希瓦氏菌包括厦门希瓦氏菌、海藻希瓦氏菌和腐败希瓦氏菌。
希瓦氏菌染色体固有携带blaOXA-48-like基因,在报道的OXA-48-like酶中有26个与希瓦氏菌固有携带的OXA高度同源,说明希瓦氏菌属可能是OXA-48-like酶的起源,并在blaOXA-48基因的传播中发挥重要作用[54]。Poirel等[55]首次在奥奈达湖希瓦氏菌中发现染色体定位的blaOXA-54基因,OXA-54与OXA-48同源性为92%,可以显著水解苯唑西林,常见的酶抑制剂仅对其有弱抑制作用。厦门希瓦氏菌与奥奈达湖希瓦氏菌亲缘关系接近,分离自我国福建厦门近海沉积物中。Dabos等[56]和Jousset等[57]在厦门希瓦氏菌中报道了染色体固有携带blaOXA-535,OXA-535与OXA-48同源性为91.3%,对多种β-内酰胺类药物有高效水解作用。在沼泽植物[54]、河水[58-59]、淡水[60]、养殖场废水[61-62]以及医院污水[63-64]等环境来源的厦门希瓦氏菌中不断检出OXA-48-like酶相关基因。因此,希瓦氏菌被认为是自然环境中耐药基因的重要载体和储存库,通过转座子介导耐药基因快速转移和传播,对人类及动物的健康造成潜在威胁。
5 总结与展望OXA酶是革兰阴性菌中种类繁多、分布广泛的一类β-内酰胺酶,可由质粒、转座子等多种可移动元件携带并转移,实现OXA酶在不同菌株间的快速传播。近年来,可以水解碳青霉烯类抗生素的OXA酶新变体不断出现,并在人群、动物及环境中传播,对公共卫生造成潜在的、巨大的威胁[10]。意大利、西班牙等南欧国家产OXA-48-like酶耐碳青霉烯类肠杆菌(carbapenem-resistant Enterobacterales, CRE)占比超过产KPC、NDM等碳青霉烯酶CRE,是CRE的主要耐药机制[65]。在我国,尽管具有碳青霉烯类药物水解功能的OXA酶在CRE中占比较低,但不断有OXA-181、OXA-232、OXA-427、OXA-484、OXA-546、OXA-894和OXA-913等新的具有碳青霉烯类药物水解活性的亚型被检出,提示需高度关注OXA酶的流行和进化。环境、动物及人群中blaOXA的检测、OXA酶功能研究和持续监测,对掌握OXA酶的分布特征和流行趋势,保障食品安全,减慢或抑制耐药基因在菌株间的传播和扩散,最大限度减少耐药菌的危害具有重要意义。
利益冲突:所有作者均声明不存在利益冲突。
| [1] |
Bonomo RA. β-lactamases: a focus on current challenges[J]. Cold Spring Harb Perspect Med, 2017, 7(1): a025239. DOI:10.1101/cshperspect.a025239 |
| [2] |
Nordmann P, Poirel L. Epidemiology and diagnostics of carbapenem resistance in Gram-negative bacteria[J]. Clin Infect Dis, 2019, 69(Suppl 7): S521-S528. |
| [3] |
Bush K. Past and present perspectives on β-lactamases[J]. Antimicrob Agents Chemother, 2018, 62(10): e01076-18. |
| [4] |
Yoon EJ, Jeong SH. Class D β-lactamases[J]. J Antimicrob Chemother, 2021, 76(4): 836-864. DOI:10.1093/jac/dkaa513 |
| [5] |
Evans BA, Amyes SGB. OXA β-lactamases[J]. Clin Micro-biol Rev, 2014, 27(2): 241-263. DOI:10.1128/CMR.00117-13 |
| [6] |
Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm![J]. Trends Mol Med, 2012, 18(5): 263-272. DOI:10.1016/j.molmed.2012.03.003 |
| [7] |
Sykes RB, Matthew M. The beta-lactamases of Gram-negative bacteria and their role in resistance to beta-lactam antibiotics[J]. J Antimicrob Chemother, 1976, 2(2): 115-157. DOI:10.1093/jac/2.2.115 |
| [8] |
Musila L, Kyany'a C, Maybank R, et al. Detection of diverse carbapenem and multidrug resistance genes and high-risk strain types among carbapenem non-susceptible clinical isolates of target Gram-negative bacteria in Kenya[J]. PLoS One, 2021, 16(2): e0246937. DOI:10.1371/journal.pone.0246937 |
| [9] |
Moghnieh RA, Kanafani ZA, Tabaja HZ, et al. Epidemiology of common resistant bacterial pathogens in the countries of the Arab League[J]. Lancet Infect Dis, 2018, 18(12): e379-e394. DOI:10.1016/S1473-3099(18)30414-6 |
| [10] |
Pandey D, Singhal N, Kumar M. Investigating the OXA variants of ESKAPE pathogens[J]. Antibiotics (Basel), 2021, 10(12): 1539. DOI:10.3390/antibiotics10121539 |
| [11] |
Abouelfetouh A, Mattock J, Turner D, et al. Diversity of carbapenem-resistant Acinetobacter baumannii and bacteriophage-mediated spread of the Oxa23 carbapenemase[J]. Microb Ge-nom, 2022, 8(2): 000752. |
| [12] |
Tafaj S, Kostyanev T, Xavier BB, et al. Clonal transmission of multidrug-resistant Acinetobacter baumannii harbouring blaOXA-24-like and blaOXA-23-like genes in a tertiary hospital in Albania[J]. J Glob Antimicrob Resist, 2020, 23: 79-81. DOI:10.1016/j.jgar.2020.07.026 |
| [13] |
Lukovic B, Gajic I, Dimkic I, et al. The first nationwide multicenter study of Acinetobacter baumannii recovered in Serbia: emergence of OXA-72, OXA-23 and NDM-1-producing isolates[J]. Antimicrob Resist Infect Control, 2020, 9(1): 101. DOI:10.1186/s13756-020-00769-8 |
| [14] |
Colquhoun JM, Farokhyfar M, Hutcheson AR, et al. OXA-23 β-lactamase overexpression in Acinetobacter baumannii drives physiological changes resulting in new genetic vulnerabilities[J]. mBio, 2021, 12(6): e0313721. DOI:10.1128/mBio.03137-21 |
| [15] |
Wang TH, Leu YS, Wang NY, et al. Prevalence of different carbapenemase genes among carbapenem-resistant Acinetobacter baumannii blood isolates in Taiwan[J]. Antimicrob Resist Infect Control, 2018, 7: 123. DOI:10.1186/s13756-018-0410-5 |
| [16] |
Rouhi S, Ramazanzadeh R. Prevalence of blaoxacillinase-23 and blaoxacillinase-24/40-type carbapenemases in Pseudomonas aeruginosa species isolated from patients with nosocomial and non-nosocomial infections in the west of Iran[J]. Iran J Pathol, 2018, 13(3): 348-356. |
| [17] |
Hujer AM, Hujer KM, Leonard DA, et al. A comprehensive and contemporary "snapshot" of β-lactamases in carbapenem resistant Acinetobacter baumannii[J]. Diagn Microbiol Infect Dis, 2021, 99(2): 115242. DOI:10.1016/j.diagmicrobio.2020.115242 |
| [18] |
Nguyen AT, Pham SC, Ly AK, et al. Overexpression of blaOXA-58 gene driven by ISAba3 is associated with imipenem resistance in a clinical Acinetobacter baumannii isolate from Vie- tnam[J]. Biomed Res Int, 2020, 2020: 7213429. |
| [19] |
Matos AP, Cayô R, Almeida LGP, et al. Genetic characteri-zation of plasmid-borne blaOXA-58 in distinct Acinetobacter species[J]. mSphere, 2019, 4(5): e00376-19. |
| [20] |
Fávaro LDS, de Paula-Petroli SB, Romanin P, et al. Detection of OXA-58-producing Acinetobacter bereziniae in Brazil[J]. J Glob Antimicrob Resist, 2019, 19: 53-55. DOI:10.1016/j.jgar.2019.08.011 |
| [21] |
Mairi A, Pantel A, Sotto A, et al. OXA-48-like carbapenemases producing Enterobacteriaceae in different niches[J]. Eur J Clin Microbiol Infect Dis, 2018, 37(4): 587-604. DOI:10.1007/s10096-017-3112-7 |
| [22] |
Boyd SE, Holmes A, Peck R, et al. OXA-48-like β-lactama-ses: global epidemiology, treatment options, and development pipeline[J]. Antimicrob Agents Chemother, 2022, 66(8): e0021622. DOI:10.1128/aac.00216-22 |
| [23] |
Pitout JDD, Peirano G, Kock MM, et al. The global ascen-dency of OXA-48-type carbapenemases[J]. Clin Microbiol Rev, 2019, 33(1): e00102-19. |
| [24] |
Liu CC, Fang YF, Zeng Y, et al. First report of OXA-181-producing Klebsiella pneumoniae in China[J]. Infect Drug Resist, 2020, 13: 995-998. DOI:10.2147/IDR.S237793 |
| [25] |
Yu ZJ, Zhang ZR, Shi LL, et al. In silico characterization of IncX3 plasmids carrying blaOXA-181 in Enterobacterales[J]. Front Cell Infect Microbiol, 2022, 12: 988236. DOI:10.3389/fcimb.2022.988236 |
| [26] |
Jia HY, Zhang Y, Ye JZ, et al. Outbreak of multidrug-resis-tant OXA-232-producing ST15 Klebsiella pneumoniae in a teaching hospital in Wenzhou, China[J]. Infect Drug Resist, 2021, 14: 4395-4407. DOI:10.2147/IDR.S329563 |
| [27] |
Zhu ZC, Huang HF, Xu YM, et al. Emergence and genomics of OXA-232-producing Klebsiella pneumoniae in a hospital in Yancheng, China[J]. J Glob Antimicrob Resist, 2021, 26: 194-198. DOI:10.1016/j.jgar.2021.05.015 |
| [28] |
Li X, Ma W, Qin Q, et al. Nosocomial spread of OXA-232-producing Klebsiella pneumoniae ST15 in a teaching hospital, Shanghai, China[J]. BMC Microbiol, 2019, 19(1): 235. DOI:10.1186/s12866-019-1609-1 |
| [29] |
Petrova A, Feodorova Y, Miteva-Katrandzhieva T, et al. First detected OXA-50 carbapenem-resistant clinical isolates Pseudomonas aeruginosa from Bulgaria and interplay between the expression of main efflux pumps, OprD and intrinsic AmpC[J]. J Med Microbiol, 2019, 68(12): 1723-1731. DOI:10.1099/jmm.0.001106 |
| [30] |
Nitz F, de Melo BO, da Silva LCN, et al. Molecular detection of drug-resistance genes of blaOXA-23-blaOXA-51 and mcr-1 in clinical isolates of Pseudomonas aeruginosa[J]. Microorga-nisms, 2021, 9(4): 786. DOI:10.3390/microorganisms9040786 |
| [31] |
Bonnin RA, Girlich D, Jousset AB, et al. A single Proteus mirabilis lineage from human and animal sources: a hidden reservoir of OXA-23 or OXA-58 carbapenemases in Entero-bacterales[J]. Sci Rep, 2020, 10(1): 9160. DOI:10.1038/s41598-020-66161-z |
| [32] |
Kang Q, Wang X, Zhao JA, et al. Multidrug-resistant Proteus mirabilis isolates carrying blaOXA-1 and blaNDM-1 from wildlife in China: increasing public health risk[J]. Integr Zool, 2021, 16(6): 798-809. DOI:10.1111/1749-4877.12510 |
| [33] |
Ramsamy Y, Mlisana KP, Amoako DG, et al. Mobile genetic elements-mediated Enterobacterales-associated carbapenemase antibiotic resistance genes propagation between the environment and humans: a one health South African study[J]. Sci Total Environ, 2022, 806(Pt 3): 150641. |
| [34] |
Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide[J]. Clin Microbiol Infect, 2014, 20(9): 821-830. DOI:10.1111/1469-0691.12719 |
| [35] |
Bush K, Jacoby GA. Updated functional classification of beta-lactamases[J]. Antimicrob Agents Chemother, 2010, 54(3): 969-976. DOI:10.1128/AAC.01009-09 |
| [36] |
Bonnin RA, Bogaerts P, Girlich D, et al. Molecular characteri- zation of OXA-198 carbapenemase-producing Pseudomonas aeruginosa clinical isolates[J]. Antimicrob Agents Chemo-ther, 2018, 62(6): e02496-17. |
| [37] |
Kotsakis SD, Flach CF, Razavi M, et al. Characterization of the first OXA-10 natural variant with increased carbapenemase activity[J]. Antimicrob Agents Chemother, 2019, 63(1): e01817-18. |
| [38] |
Koirala J, Tyagi I, Guntupalli L, et al. OXA-23 and OXA-40 producing carbapenem-resistant Acinetobacter baumannii in central Illinois[J]. Diagn Microbiol Infect Dis, 2020, 97(1): 114999. DOI:10.1016/j.diagmicrobio.2020.114999 |
| [39] |
Yang ZH, Wang P, Song P, et al. Carbapenemase OXA-423: a novel OXA-23 variant in Acinetobacter baumannii[J]. Infect Drug Resist, 2020, 13: 4069-4075. DOI:10.2147/IDR.S277364 |
| [40] |
Turton JF, Ward ME, Woodford N, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii[J]. FEMS Microbiol Lett, 2006, 258(1): 72-77. DOI:10.1111/j.1574-6968.2006.00195.x |
| [41] |
Wu WY, He Y, Lu J, et al. Transition of blaOXA-58-like to blaOXA-23-like in Acinetobacter baumannii clinical isolates in southern China: an 8-year study[J]. PLoS One, 2015, 10(9): e0137174. DOI:10.1371/journal.pone.0137174 |
| [42] |
Kopotsa K, Osei Sekyere J, Mbelle NM. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review[J]. Ann N Y Acad Sci, 2019, 1457(1): 61-91. DOI:10.1111/nyas.14223 |
| [43] |
Aertker KMJ, Chan HTH, Lohans CT, et al. Analysis of β-lactone formation by clinically observed carbapenemases informs on a novel antibiotic resistance mechanism[J]. J Biol Chem, 2020, 295(49): 16604-16613. DOI:10.1074/jbc.RA120.014607 |
| [44] |
Sommer J, Gerbracht KM, Krause FF, et al. OXA-484, an OXA-48-type carbapenem-hydrolyzing class D β-lactamase from Escherichia coli[J]. Front Microbiol, 2021, 12: 660094. DOI:10.3389/fmicb.2021.660094 |
| [45] |
Dabos L, Bogaerts P, Bonnin RA, et al. Genetic and biochemi- cal characterization of OXA-519, a novel OXA-48-like β-lactamase[J]. Antimicrob Agents Chemother, 2018, 62(8): e00469-18. |
| [46] |
Dortet L, Oueslati S, Jeannot K, et al. Genetic and biochemical characterization of OXA-405, an OXA-48-type extended-spectrum β-lactamase without significant carbapenemase activity[J]. Antimicrob Agents Chemother, 2015, 59(7): 3823-3828. DOI:10.1128/AAC.05058-14 |
| [47] |
Poirel L, Castanheira M, Carrёr A, et al. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins[J]. Antimicrob Agents Chemother, 2011, 55(6): 2546-2551. DOI:10.1128/AAC.00022-11 |
| [48] |
Gomez S, Pasteran F, Faccone D, et al. Intrapatient emergence of OXA-247: a novel carbapenemase found in a patient previously infected with OXA-163-producing Klebsiella pneumoniae[J]. Clin Microbiol Infect, 2013, 19(5): E233-E235. DOI:10.1111/1469-0691.12142 |
| [49] |
Vázquez-Ucha JC, Arca-Suárez J, Bou G, et al. New carbape-nemase inhibitors: clearing the way for the β-lactams[J]. Int J Mol Sci, 2020, 21(23): 9308. DOI:10.3390/ijms21239308 |
| [50] |
Karaiskos I, Galani I, Souli M, et al. Novel β-lactam-β-lactamase inhibitor combinations: expectations for the treatment of carbapenem-resistant Gram-negative pathogens[J]. Expert Opin Drug Metab Toxicol, 2019, 15(2): 133-149. DOI:10.1080/17425255.2019.1563071 |
| [51] |
Yahav D, Giske CG, Grāmatniece A, et al. New β-lactam-β-lactamase inhibitor combinations[J]. Clin Microbiol Rev, 2020, 34(1): e00115-20. |
| [52] |
Janda JM, Abbott SL. The genus Shewanella: from the briny depths below to human pathogen[J]. Crit Rev Microbiol, 2014, 40(4): 293-312. DOI:10.3109/1040841X.2012.726209 |
| [53] |
Wang JH, Tseng SY, Tung KC. Genomic investigation of emerging zoonotic pathogen Shewanella xiamenensis[J]. Tzu Chi Med J, 2020, 32(2): 162-166. DOI:10.4103/tcmj.tcmj_69_19 |
| [54] |
Tacão M, Araújo S, Vendas M, et al. Shewanella species as the origin of blaOXA-48 genes: insights into gene diversity, associated phenotypes and possible transfer mechanisms[J]. Int J Antimicrob Agents, 2018, 51(3): 340-348. DOI:10.1016/j.ijantimicag.2017.05.014 |
| [55] |
Poirel L, Héritier C, Nordmann P. Chromosome-encoded ambler class D beta-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase[J]. Antimicrob Agents Chemother, 2004, 48(1): 348-351. DOI:10.1128/AAC.48.1.348-351.2004 |
| [56] |
Dabos L, Jousset AB, Bonnin RA, et al. Genetic and biochemical characterization of OXA-535, a distantly related OXA-48-like β-lactamase[J]. Antimicrob Agents Chemother, 2018, 62(10): e01198-18. |
| [57] |
Jousset AB, Dabos L, Bonnin RA, et al. CTX-M-15-producing Shewanella species clinical isolate expressing OXA-535, a chromosome-encoded OXA-48 variant, putative progenitor of the plasmid-encoded OXA-436[J]. Antimicrob Agents Chemother, 2018, 62(1): e01879-17. |
| [58] |
Tafoukt R, Leangapichart T, Hadjadj L, et al. Characterisa-tion of blaOXA-538, a new variant of blaOXA-48, in Shewanella xiamenensis isolated from river water in Algeria[J]. J Glob Antimicrob Resist, 2018, 13: 70-73. DOI:10.1016/j.jgar.2017.11.008 |
| [59] |
Tacão M, Correia A, Henriques I. Environmental Shewanella xiamenensis strains that carry blaOXA-48 or blaOXA-204 genes: additional proof for blaOXA-48-like gene origin[J]. Antimicrob Agents Chemother, 2013, 57(12): 6399-6400. DOI:10.1128/AAC.00771-13 |
| [60] |
Ceccarelli D, van Essen-Zandbergen A, Veldman KT, et al. Chromosome-based blaOXA-48-like variants in Shewanella species isolates from food-producing animals, fish, and the aquatic environment[J]. Antimicrob Agents Chemother, 2017, 61(2): e01013-16. |
| [61] |
Zou HY, Zhou ZY, Xia HY, et al. Characterization of chromosome-mediated BlaOXA-894 in Shewanella xiamenensis isolated from pig wastewater[J]. Int J Environ Res Public Health, 2019, 16(19): 3768. DOI:10.3390/ijerph16193768 |
| [62] |
Irrgang A, Pauly N, Tenhagen BA, et al. Spill-over from public health? First detection of an OXA-48-producing Escherichia coli in a German pig farm[J]. Microorganisms, 2020, 8(6): 855. DOI:10.3390/microorganisms8060855 |
| [63] |
Yousfi K, Touati A, Lefebvre B, et al. A novel plasmid, pSx1, harboring a new Tn1696 derivative from extensively drug-resistant Shewanella xiamenensis encoding OXA-416[J]. Microb Drug Resist, 2017, 23(4): 429-436. DOI:10.1089/mdr.2016.0025 |
| [64] |
Barraud O, Casellas M, Dagot C, et al. An antibiotic-resistant class 3 integron in an Enterobacter cloacae isolate from hospital effluent[J]. Clin Microbiol Infect, 2013, 19(7): E306-E308. DOI:10.1111/1469-0691.12186 |
| [65] |
Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace[J]. J Antimicrob Chemother, 2012, 67(7): 1597-1606. DOI:10.1093/jac/dks121 |



